AIM:
To find the protein families of specific pattern by using PHI BLAST.
INTRODUCTION:
PHI-BLAST (Pattern-Hit Initiated BLAST) is a search program that combines matching of regular expressions with local alignments surrounding the match. The most important features of the program have been incorporated into the BLAST software framework partly for user convenience and partly so that PHI-BLAST may be combined seamlessly with PSI-BLAST. Other features that do not fit into the BLAST framework will be released later as a separate program and/or separate webpage query options.
One very restrictive way to identify protein motifs is by regular expressions that must contain each instance of the motif. The PROSITE database is a compilation of restricted regular expressions that describe protein motifs. Given a protein sequence S and a regular expression pattern P occurring in S, PHI-BLAST helps answer the question: What other protein sequences both contain an occurrence of P and are homologous to S in the vicinity of the pattern occurrences? PHI-BLAST may be preferable to just searching for pattern occurrences because it filters out those cases where the pattern occurrence is probably random and not indicative of homology. PHI-BLAST may be preferable to other flavours of BLAST because it is faster and because it allows the user to express a rigid pattern occurrence requirement.
PROCEDURE:
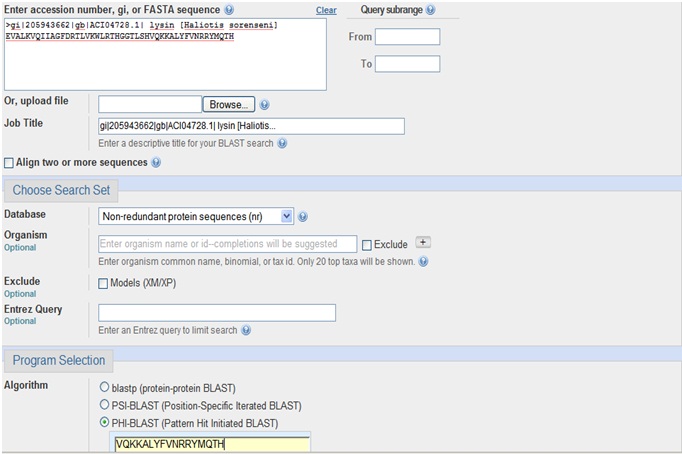
- Obtain sequences of given accession no. from NCBI in FASTA format
- Enter the sequences into the clustalW software in the given space
- Obtain pattern of given sequences
- Open the PHI BLAST program
- Paste given query sequence and pattern of it
- Click run button to perform search operation




No comments:
Post a Comment